A central tenet in the Abouheif lab is to always keep an open mind. Prof. Ehab Abouheif always likes to say that “ultimately data talks.” This project has been a passion of ours for the last 5 years. The ‘Blochmannia project’ as it became known, was born out of the convergence of multiple independent, seemingly unrelated projects. Upon reflection, our journey was filled with serendipitous twists and turns, so much so, that it would have been impossible to predict where we ended up from where we began.

In 2013, the Abouheif lab was in the midst of establishing a developmental table for the Florida carpenter ant, Camponotus floridanus, – from the development of a freshly laid egg through metamorphosis to the adult. Ehab’s vision for this table was to champion the eco-evo-devo program by building developmental tools for the social insect community. While characterizing the morphology of C. floridanus embryos, we discovered a large cloud at the posterior pole that accumulated large amounts of a nuclear dye called DAPI. To determine the composition of this DAPI-positive cloud, we injected C. floridanus embryos with enzymes that degrade different macromolecules. After treating embryos with RNase and Xylenes, which degrade RNAs and lipids, we found that only DNase was capable of eliminating this DAPI-positive cloud from the posterior pole. We therefore concluded the cloud contained DNA. However, its purpose would remain a mystery to us for over a year.

Left: : Freshly laid C. floridanus embryo with DAPI-positive mass at the posterior pole. Right: Colony of C. floridanus.
During this period, Matteen – then a postdoc in the lab – was studying the body plan morphology of ants, especially the presence of the highly constricted waist that entomologists call the ‘petiole.’ The petiole is generated through the modification of the second and third abdominal segment, which during development are known to be under the control of the Hox genes abdominal A (abdA) and Ultrabithorax (Ubx). To understand the role of these Hox genes in petiole evolution in ants, Matteen knocked down abdA and Ubx using RNA-interference. Instead of finding changes in segment identity, which occur in all other insects, he discovered that the embryos were truncated, where only the most anterior segments remained. It would take 2 years to accept and make sense of this phenotype.


surrounding a void. Right: Void in ctyoplasm is where
germplasm is localized
I joined the lab in 2014. My thesis proposal was to follow up on the work of former post-doc Abderrahman Khila (Abdou), to understand the molecular basis of reproduction in ants. A hallmark of ant societies is a reproductive division of labor between queen and worker castes. Queens perform almost all reproductive functions in the colony, while workers have a reduced capacity to reproduce. Abdou had discovered that this reduced capacity is caused by developmental interruptions in worker reproduction. The early phases of my work would entail cloning and testing antibodies for multiple germline genes to characterize germline formation of C. floridanus. I started characterizing germplasm, a region of cytoplasm that functions to specify the germline and abdomen. I found that, similar to embryos, the cloud of DNA was localized at the posterior pole of C. floridanus oocytes and surrounded its germplasm, suggesting the DNA was somehow attracted to the germline. The presence of this cloud in both oocytes and embryos made clear to us that the DNA material was being inherited from mother to daughter. This finding prompted Matteen and I to ask what this cloud of DNA is and what it is doing to the ant’s germline.

a novel mode of genetic inheritance!!!...
We then analyzed the cloud of DNA using confocal microscopy and found it was strikingly similar to polytene-like chromosomes. Therefore, based on its interaction with germplasm, we believed we had uncovered a novel mode of genetic inheritance that was both non-nuclear and non-mitochondrial. In parallel, we also found that abdA and Ubx were co-localized with the maternal DNA and expressed much earlier in development than in other insects. Together, these results suggested that somehow this novel mode of genetic transmission involved the Hox genes! Based on these findings, Matteen presented a poster on this work in Paris. A conference that would upend our research lives as we knew it. The conference went great, many thought this new maternal DNA was fascinating. Prof. Roy Gross and Prof. Heike Feldhaar, who are experts in ant microbiomes, attended the conference and saw the poster and wanted to Skype about our findings. Within 5 minutes of that meeting, it became clear that the cloud of DNA, which we had thought we discovered, was actually an obligate endosymbiont called Blochmannia (it’s official name is Candidatus blochmannia floridanus). And on top of it, it was not just any endosymbiont, but actually one of the first animal obligate endosymbionts ever described by Friedrich Blochmann in 1892.

This was humbling to say the least – to have accidentally stumbled onto an existing research program while thinking it was new. At this point we had two options, cut our losses and go back to our independent projects or persist. We decided to take this blow in stride. We kept an open mind and knew somewhere deep down there was still something interesting to be explored here. Starting from scratch, we proceeded to probe what our existing data could mean. While the co-localization of the C. floridanus germplasm and Blochmannia made sense in light of the fact that Blochmannia is vertically transmitted, we could not wrap our heads around the co-localization of Hox genes abdA and Ubx with Blochmannia. This made us wonder whether Blochmannia had anything to do with our enigmatic truncation phenotypes.
In between existential crises stemming from the fiasco of our proposed ‘the novel mode of inheritance’, I decided to continue my work on the germline of ants. I cloned a number of other germline genes, including oskar, nanos, and staufen, and used C. floridanus embryos as a positive control to test if they mark the germ cells. This single in situ was a game changer. We found that unlike in flies where these genes are expressed altogether in a single location at the posterior pole, in C. floridanus, oskar, nanos and staufen marked 3 zones of germline localization and expression, in different combinations. Our eureka moment was the realization that Hox gene and germline gene expression co-localized in these 3 zones! Our follow-up experiments showed that the Hox genes abdA and Ubx were actually upstream of these germline genes. This was very unexpected, as it suggested that the classic and highly conserved segmentation hierarchy had been turned downside up! To add the eco to our devo findings we next tested the role of Blochmannia in regulating these 3 zones of Hox gene and germline genes, by feeding C. floridanus colonies with rifampicin, an antibiotic known to eliminate Blochmannia. We discovered that Blochmannia both maintains and selectively regulates the localization and expression of key Hox and germline genes!

Bacteriocytes –Z1b and from the novel germline – Z2 (bottom right)

We next set out to put the evo in our now eco-devo findings. Obligate endosymbiosis is considered to be a major evolutionary transition in individuality, where two independently replicating taxa developmentally integrate to become a singly replicating individual. Such major evolutionary transitions in individuality are thought to be crucial for increases in biological complexity. Therefore, we started to investigate the evolutionary origins and elaboration of the developmental integration underlying the obligate endosymbiosis between Blochmannia and the ant tribe Camponotini, which C. floridanus belongs to. By chance we already had 5 other Camponotus species in the lab, which were ‘randoms’ from our various collecting trips (and just by sheer luck their phylogenetic relationships were previously described!). The lab then traveled to Florida, and with the expertise of myrmecologist Lloyd Davis, roamed the swamps to collect a species from the genus Colobopsis, which is a early branching species within the Camponotini. This would help us reconstruct how Hox and germline genes were expressed close to the origin of the obligate endosymbiosis. Finally, we chose Lasius niger, a closely related ant without an obligate endosymbiont as an outgroup. We discovered that the 3 zones of Hox and germline expression were the result of two successive steps of duplication of zones: Lasius niger had no early Hox gene and only 1 zone of germline gene expression; Colobopsis at the base of the tribe, had evolved two zones of Hox and germline; C. floridanus and more derived Camponotus species had 3 zones of Hox and germline. Furthermore, Hox genes had evolved maternal localization in Colobopsis. In the end, we concluded that the evolution of the endosymbiosis between Blochmannia and Camponotini caused all of the radical changes in early development to integrate into the Camponotini.

Left: Colobopsis embryo with two zones of germline gene expression. Right: C. floridanus embryo with 3 zones of germline gene expression (marked by nanos mRNA).
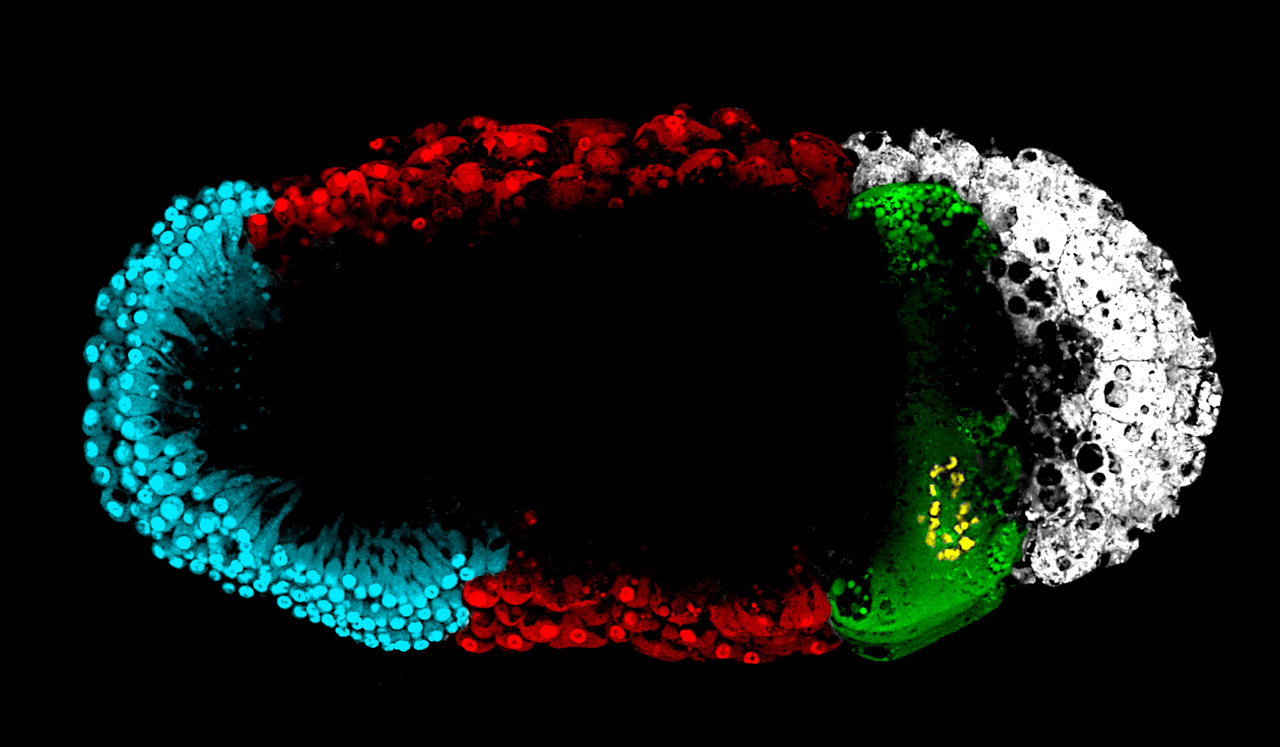
After 3 years we were ready to submit the paper and held our breath. Following our celebration that it was sent out for review; we got the reviews back. Responding to all three reviews had dramatically improved our manuscript. In our initial submission to study germline genes we visualized the localization and expression of osk, nanos and staufen mRNA, however, we had not studied what many consider to be a universal germline marker – Vasa. The reason for this was that, for the life of me, I could not get any immunohistochemistry protocols to work on embryos, which was why I focused my efforts on mRNA probes in the first place. But as the classic saying goes “necessity is the mother of invention” and after toiling away for what seemed like forever, we got Vasa to work. After Vasa, other key germline genes such as Aubergine, Tudor, Germcell-less and HSP90 began to work. To our surprise, we found localization and expression of these proteins in not three but four locations! Then like the Baader-Meinhof effect, we suddenly started seeing the fourth localization zone everywhere! Going back through our old data we ended up finding this pattern for genes we had already analyzed but had not picked up on due to its subtle expression pattern.

Four zones of localization of germline and Hox genes
Furthermore, in our initial submission our outgroup to trace the origin of the changes induced by the evolution of obligate endosymbiosis in the Camponitini was the common garden ant Lasius niger. Although within ants, Lasius is a closely related genus to Camponotus, at higher taxonomic resolution, Lasius is more like a cousin rather than a sister, a fact pointed out by our reviewers. This raised the possibility that the changes in early development we observed in the Camponotini cannot be concluded definitively to be driven by Blochmannia and that looking at other genera that evolved between Lasius and Camponotus would either validate or further nuance our findings. We did not know this at the time, but just like with Lays chips where “you can’t eat just one,” each species we looked at forced us to look at more. We started with 8 but ended up with 31 species! One of the closest related genera to the Camponitini is Gigantiops. I was surprised to find that, instead of a single zone of germline gene expression, we found two! As it turned out the appearance of a novel zone of germline and early Hox gene expression prior to the origin of the obligate endosymbiosis between Blochmannia and Camponitni was just tip of the iceberg! In addition to Gigantiops we found an additional localization zone of germline and Hox genes in multiple lineages that are sister and cousin taxa to the Camponotini. This increase in sampling enabled us to perform a formal ancestral state reconstruction. A huge thank you to Angelly Vasquez Correa, a fellow Abouheif lab PhD student for her help!

Left: Original sampling. Right: Revised sampling. Reconstruction of 5 developmental characters across 31 species as indicated by the symbols at the tips of the branches in the tree
Altogether, our results showed the existence of pre-existing developmental capacities that were essential for Blochmannia to developmentally integrate into the Camponitini. We predict that similar pre-existing developmental capacities in core developmental networks are also essential for other major transitions in evolution, whether for the evolution of a soma in multicellular organisms or the worker caste in eusocial insect societies. With this behind the scenes, I hope to have shared with you the twists and turns of our scientific journey – science is messy but it’s exciting! As assuring it is to plan, it’s equally important to leave room for surprises and side projects! Hypotheses can be wrong but can be refined or changed completely. The ability to maintain a pluralistic view, and to not hold any hypothesis as truth, facilitates a flexibility and robustness in research programs and opens you up to the unimaginable.
Check out our paper!!!!: Ab. Matteen Rafiqi, Arjuna Rajakumar, Ehab Abouheif. Origin and elaboration of a major evolutionary transition in individuality. Nature. 2020. https://www.nature.com/articles/s41586-020-2653-6






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in